Fitting Likelihood Maps
There are circumstances where you might have a 2D map of planet likelihood vs. position in the plane of the sky ($\Delta$ R.A. and Dec.). These could originate from:
- cleaned interferometer data
- some kind of spectroscopic cube model fitting to detect planets
- some other imaginative modelling process I haven't thought of!
You can feed such 2D likelihood maps in to Octofitter. Simply pass in a list of maps and a platescale mapping the pixel size to a separation in milliarcseconds. You can of course also mix these likelihood maps with relative astrometry, radial velocity, proper motion, images, etc.
If your likelihood map is not centered on the star, you can specify offset dimensions as shown below.
Image modelling is supported in Octofitter via the extension package OctofitterImages. To install it, run pkg> add http://github.com/sefffal/Octofitter.jl:OctofitterImages
For simple models of interferometer data, OctofitterInterferometry.jl can already handle fitting point sources directly to visibilities.
using Octofitter
using Distributions
using OctofitterImages
using Pigeons
using AstroImagesTypically one would load your likelihood maps from eg. FITS files like so:
# If centred at the star:
image1 = AstroImages.recenter(AstroImages.load("image-example-1.fits",1))
# If not centered at the star:
image1 = AstroImages.load("image-example-1.fits")
image1_offset = AstroImage(
image1,
# Specify coordinates here:
(
# X coordinates should go from negative to positive.
# The image should have +RA at the left.
X(-4.85878653527304:1.0:95.14121346472696),
Y(-69.0877222942365:1.0:30.9122777057635)
)
# Below, there is a platescale option. `platescale` multiplies
# these values by a scaling factor. It can be 1 if the coordinates
# above are already in milliarcseconds.
)
imview(image1_offset)If you're using a FITS file, make sure to store your data in 64-bit floating point format.
For this demonstration, however, we will construct two synthetic likelihood maps using a template orbit. We will create three peaks in two epochs.
orbit_template = orbit(
a = 1.0,
e = 0.1,
i = 0.0,
ω = 0.0,
Ω = 0.5,
plx = 50.0,
M = 1
)
epochs = [
mjd("2024-01-30"),
mjd("2024-02-29"),
]
## Create simulated data with three likelihood peaks at both epochs
x1,y1 = raoff(orbit_template,epochs[1]), decoff(orbit_template,epochs[1])
# The three peaks in our likelihood map
d1 = MvNormal([x1, y1], [
5.0 0.2
0.2 8.0
])
d2 = MvNormal([x1+8.0,y1+4], [
5.0 0.6
0.6 5.0
])
d3 = MvNormal([x1+9.0, y1-10.0], [
6.0 0.6
0.6 6.0
])
d = MixtureModel([d1, d2, d3], [0.5, 0.3, 0.2])
# Calculate a log-likelihood map over a +-50 mas patch around (x1, x2)
lm1 = broadcast(x1 .+ (-50:50), y1 .+ (-50:1:50)') do x, y
logpdf(d, [x, y])
end
# Place in an AstroImage with appropriate offset coordinates
image1_offset = AstroImage(
lm1,
# Specify coordinates here:
(
# X coordinates should go from negative to positive.
# The image should have +RA at the left.
X(x1 .+ (-50:50)),
Y(y1 .+ (-50:1:50))
)
# Below, there is a platescale option. `platescale` multiplies
# these values by a scaling factor. It can be 1 if the coordinates
# above are already in milliarcseconds.
)
imview(10 .^ image1_offset)That was the first epoch. We now generate data for the second epoch:
x2,y2 = raoff(orbit_template,epochs[2]), decoff(orbit_template,epochs[2])
# The three peaks in our likelihood map
d1 = MvNormal([x2, y2], [
5.0 0.2
0.2 8.0
])
d2 = MvNormal([x2+10.0,y2], [
5.0 0.6
0.6 5.0
])
d3 = MvNormal([x2-4.0, y2-10.0], [
6.0 0.6
0.6 6.0
])
d = MixtureModel([d1, d2, d3], [0.5, 0.3, 0.2])
lm2 = broadcast(x2 .+ (-50:50), y2 .+ (-50:1:50)') do x, y
logpdf(d, [x, y])
end
# Place in an AstroImage with appropriate offset coordinates
image2_offset = AstroImage(
lm2,
# Specify coordinates here:
(
# X coordinates should go from negative to positive.
# The image should have +RA at the left.
X(x2 .+ (-50:50)),
Y(y2 .+ (-50:1:50))
)
# Below, there is a platescale option. `platescale` multiplies
# these values by a scaling factor. It can be 1 if the coordinates
# above are already in milliarcseconds.
)
imview(10 .^ image2_offset)Okay, we have our synthetic data. We now set up a LogLikelihoodMap object to contain our matrices of log likelihood values:
First, create a table of our likelihood map observations:
likemap_dat = Table(;
epoch = epochs,
map = [image1_offset, image2_offset],
platescale = [1.0, 1.0] # milliarcseconds/pixel of the map
)
loglikemap = LogLikelihoodMap(
likemap_dat,
name="GRAVITY",
variables=@variables begin
platescale = 1.0 # Platescale multiplier [could use: platescale ~ truncated(Normal(1, 0.01), lower=0)]
northangle = 0.0 # North angle offset in radians [could use: northangle ~ Normal(0, deg2rad(1))]
end
);OctofitterImages.LogLikelihoodMap Table with 5 columns and 2 rows:
epoch map platescale fillvalue mapinterp
┌───────────────────────────────────────────────────────────────────────────
1 │ 60339.0 [-393.192 -387.534 … 1.0 -423.544 101×101 extrapolate…
2 │ 60369.0 [-287.052 -281.177 … 1.0 -423.544 101×101 extrapolate…We now create a one-planet model and run the fit using octofit_pigeons. This parallel-tempered sampler is slower than the regular octofit, but is recommended over the default Hamiltonian Monte Carlo sampler due to the multi-modal nature of the data.
planet_b = Planet(
name="b",
basis=Visual{KepOrbit},
likelihoods=[loglikemap],
variables=@variables begin
a ~ Uniform(0, 10)
e ~ Uniform(0.0, 0.5)
i ~ Sine()
M = system.M
ω ~ UniformCircular()
Ω ~ UniformCircular()
θ ~ UniformCircular()
tp = θ_at_epoch_to_tperi(θ, 60339.0; M, e, a, i, ω, Ω) # reference epoch for θ. Choose an MJD date near your data.
end
)
sys = System(
name="Tutoria",
companions=[planet_b],
likelihoods=[],
variables=@variables begin
M ~ truncated(Normal(1.0, 0.1), lower=0.1)
plx ~ truncated(Normal(50.0, 0.02), lower=0.1)
end
)
model = Octofitter.LogDensityModel(sys)
chain, pt = octofit_pigeons(model, n_rounds=10) # increase n_rounds until log(Z₁/Z₀) converges.
display(chain)[ Info: Preparing model
┌ Info: Determined number of free variables
└ D = 11
┌ Info: Determined number type
└ T = Float64
ℓπcallback(θ): 0.000015 seconds
∇ℓπcallback(θ): 0.000023 seconds (1 allocation: 32 bytes)
┌ Warning: This model has priors that cannot be sampled IID.
└ @ OctofitterPigeonsExt ~/work/Octofitter.jl/Octofitter.jl/ext/OctofitterPigeonsExt/OctofitterPigeonsExt.jl:95
[ Info: Sampler running with multiple threads : true
[ Info: Likelihood evaluated with multiple threads: false
[ Info: Performing global optimization with 11 parameters (0 initial parameter value provided).
┌ Info: Found sample of initial positions
│ logpost_range = (-19.357335961743463, -8.874643268140106)
└ mean_logpost = -12.434866610440666
────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
scans restarts Λ Λ_var time(s) allc(B) log(Z₁/Z₀) min(α) mean(α) min(αₑ) mean(αₑ)
────────── ────────── ────────── ────────── ────────── ────────── ────────── ────────── ────────── ────────── ──────────
2 0 2.4 2.62 0.227 4.15e+06 -41.8 3.36e-25 0.838 1 1
4 0 3.89 4.53 0.0626 1.49e+05 -24.7 4.81e-05 0.728 1 1
8 0 3.34 4.29 0.121 2.54e+05 -18.7 0.0599 0.754 1 1
16 0 5.05 3.67 0.241 4.86e+05 -20.5 0.175 0.719 1 1
32 0 4.77 4.56 0.479 7.01e+05 -21.7 0.354 0.699 1 1
64 3 4.7 3.04 1.11 4.64e+07 -21.4 0.436 0.75 1 1
128 17 4.72 3.03 1.87 7.9e+07 -20.7 0.556 0.75 1 1
256 35 4.64 3.23 3.81 1.58e+08 -20.8 0.572 0.746 1 1
512 71 4.7 3.12 7.57 3.14e+08 -21 0.621 0.748 1 1
1.02e+03 161 4.84 3.05 15.2 6.27e+08 -21.1 0.645 0.745 1 1
────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────octofit_pigeons scales very well across multiple cores. Start julia with julia --threads=auto to make sure you have multiple threads available for sampling.
Display the results:
using CairoMakie
octoplot(model, chain)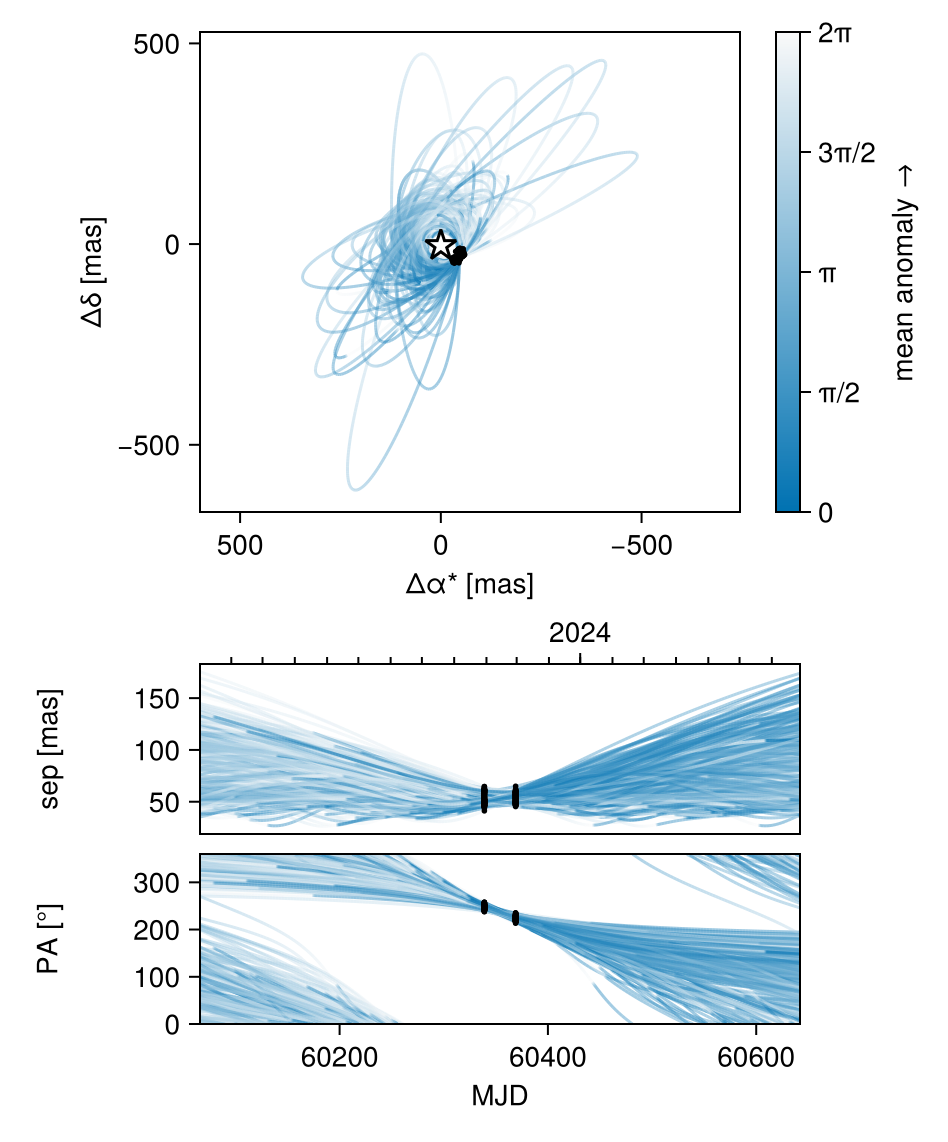
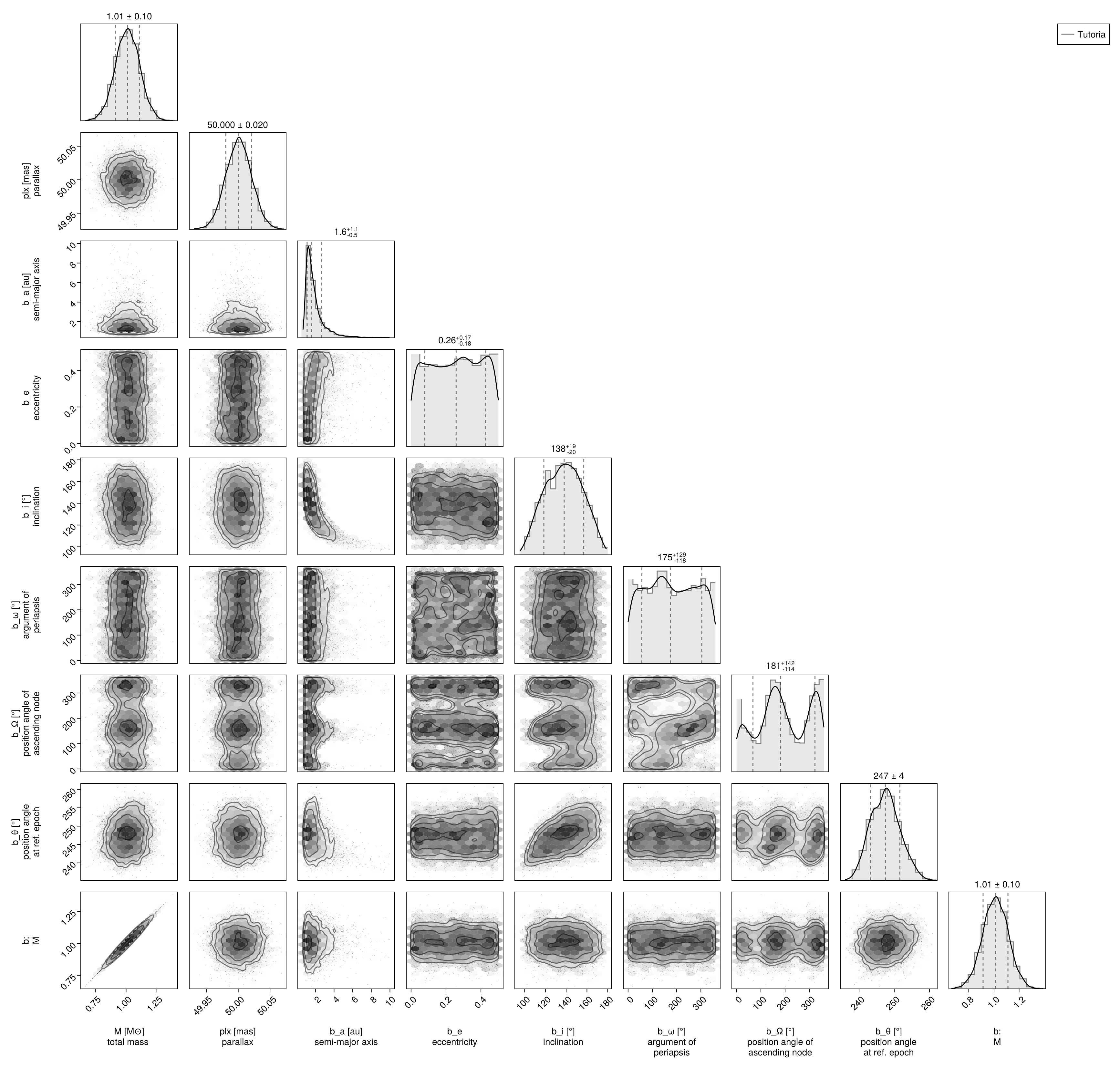
Corner plot:
using CairoMakie, PairPlots
octocorner(model,chain,small=true,)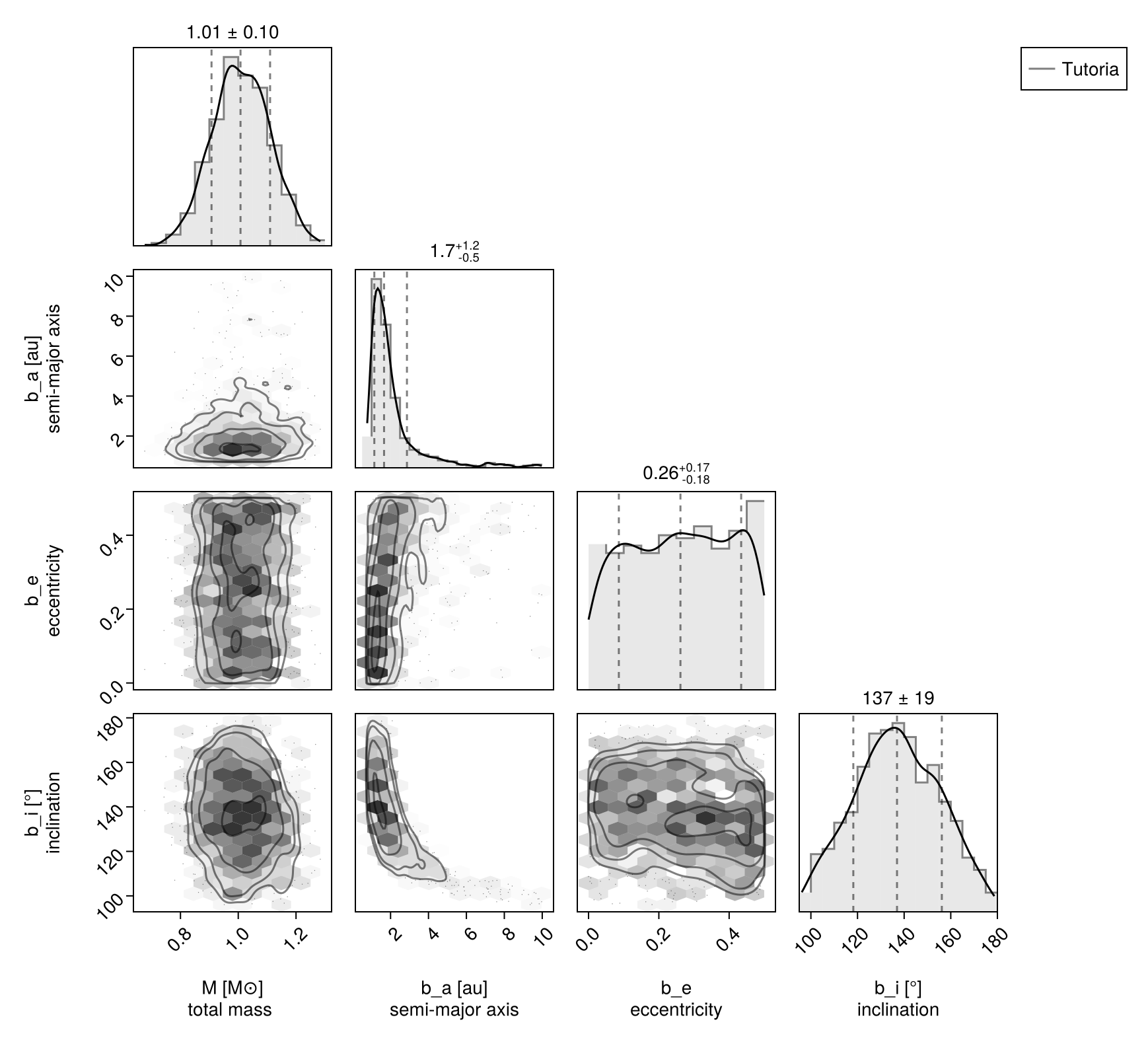
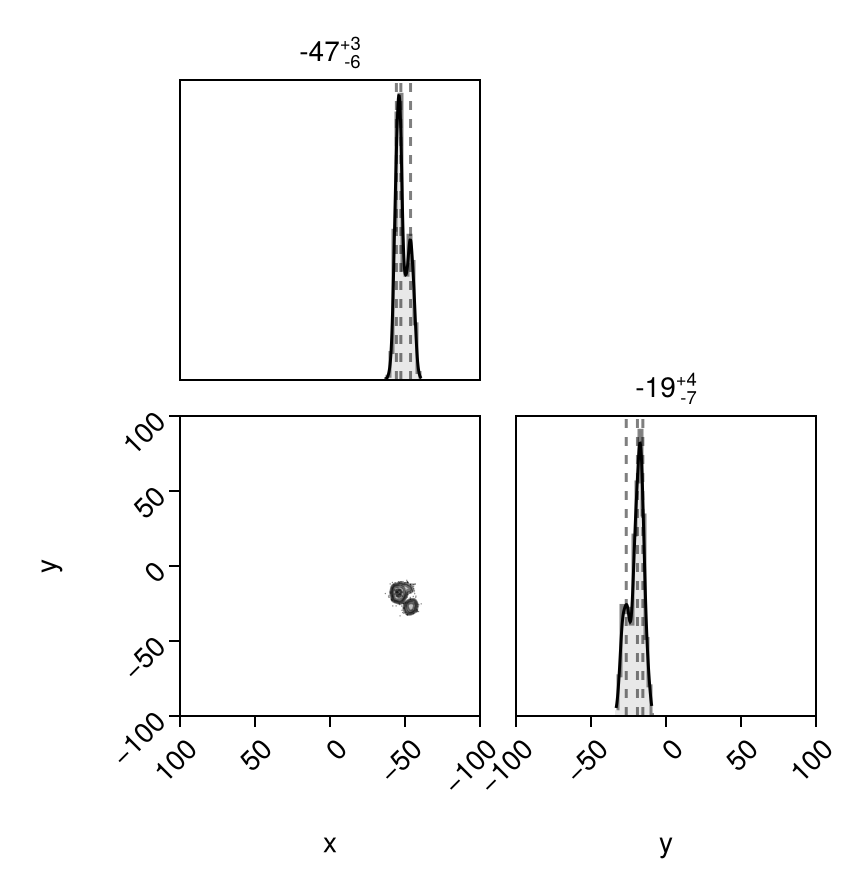
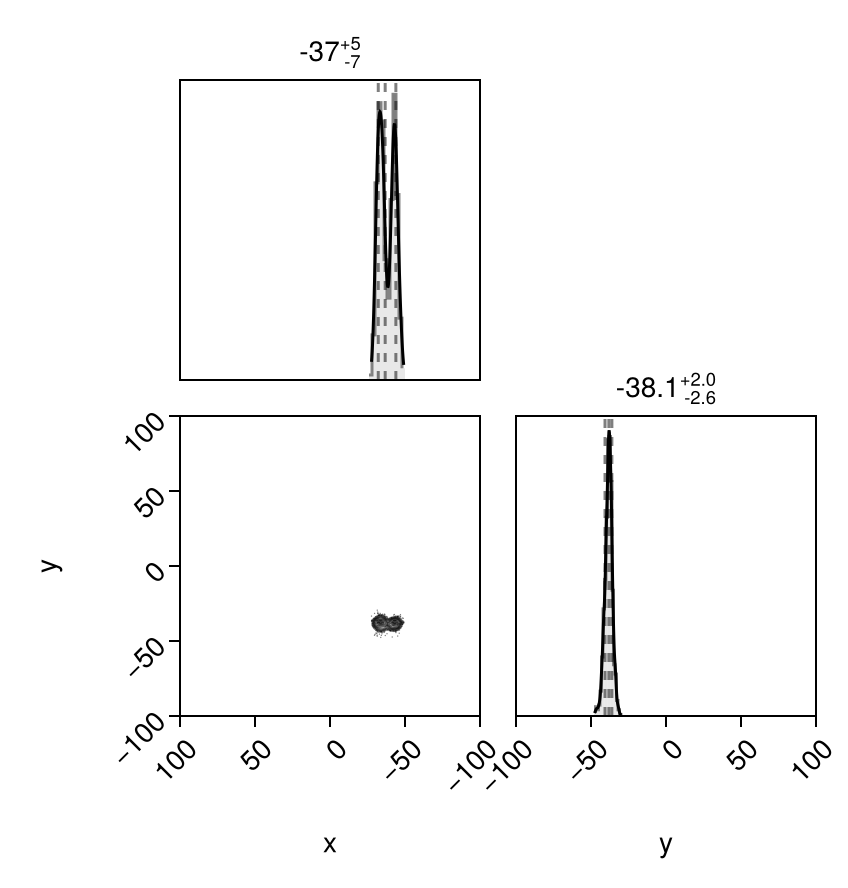
And finally let's look at the posterior predictive distributions at both epochs:
els = Octofitter.construct_elements(model, chain,:b, :)
x = raoff.(els, loglikemap.table.epoch[1])
y = decoff.(els, loglikemap.table.epoch[1])
pairplot(
(;x,y),
axis=(
x = (;
lims=(low=100,high=-100)
),
y = (;
lims=(low=-100,high=100)
)
)
)
x = raoff.(els, loglikemap.table.epoch[2])
y = decoff.(els, loglikemap.table.epoch[2])
pairplot(
(;x,y),
axis=(
x = (;
lims=(low=100,high=-100)
),
y = (;
lims=(low=-100,high=100)
)
)
)
Resume sampling for additional rounds
If you would like to add additional rounds of sampling, you may do the following:
pt = increment_n_rounds!(pt, 2)
chain, pt = octofit_pigeons(pt)(chain = MCMC chain (4096×22×1 Array{Float64, 3}), pt = PT(checkpoint = false, ...))Updated corner plot:
using CairoMakie, PairPlots
octocorner(model,chain,small=false,)